Alignment of Communities with Different Functional Types
ex2-func_type.Rmd
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(fsmc)
library(magrittr)
library(purrr)
#>
#> Attaching package: 'purrr'
#> The following object is masked from 'package:magrittr':
#>
#> set_names
library(readr)
library(stringr)
library(tidyr)
#>
#> Attaching package: 'tidyr'
#> The following object is masked from 'package:magrittr':
#>
#> extractselection <- c(“A1R12”, “B3M02”, “3D05”) facilitation <- c(“E3M17”, “D2R05”, “E3R18”)
Load Data
ft_url <- "https://raw.githubusercontent.com/admarhi/fsmc-data/main/misosoup/func_type.csv"
ft_tb <-
read_csv(ft_url) %>%
group_by(func_type, substrate, focal_strain) %>%
filter(solution <= 2) %>%
ungroup() %>%
nest(.by = c("func_type", "substrate", "focal_strain", "solution")) %>%
mutate(name = paste(substrate, focal_strain, solution, sep = "_"))
#> Rows: 76606 Columns: 7
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> chr (5): func_type, substrate, focal_strain, species, metabolite
#> dbl (2): solution, flux
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
selection_tb <- ft_tb %>%
filter(func_type == "selection")
selection <-
set_names(selection_tb$data, selection_tb$name) %>%
imap(newMiCo)
facilitation_tb <- ft_tb %>%
filter(func_type == "facilitation")
facilitation <-
set_names(facilitation_tb$data, facilitation_tb$name) %>%
imap(newMiCo)
plotAlignmentHeatmap(selection_alig, 0.5)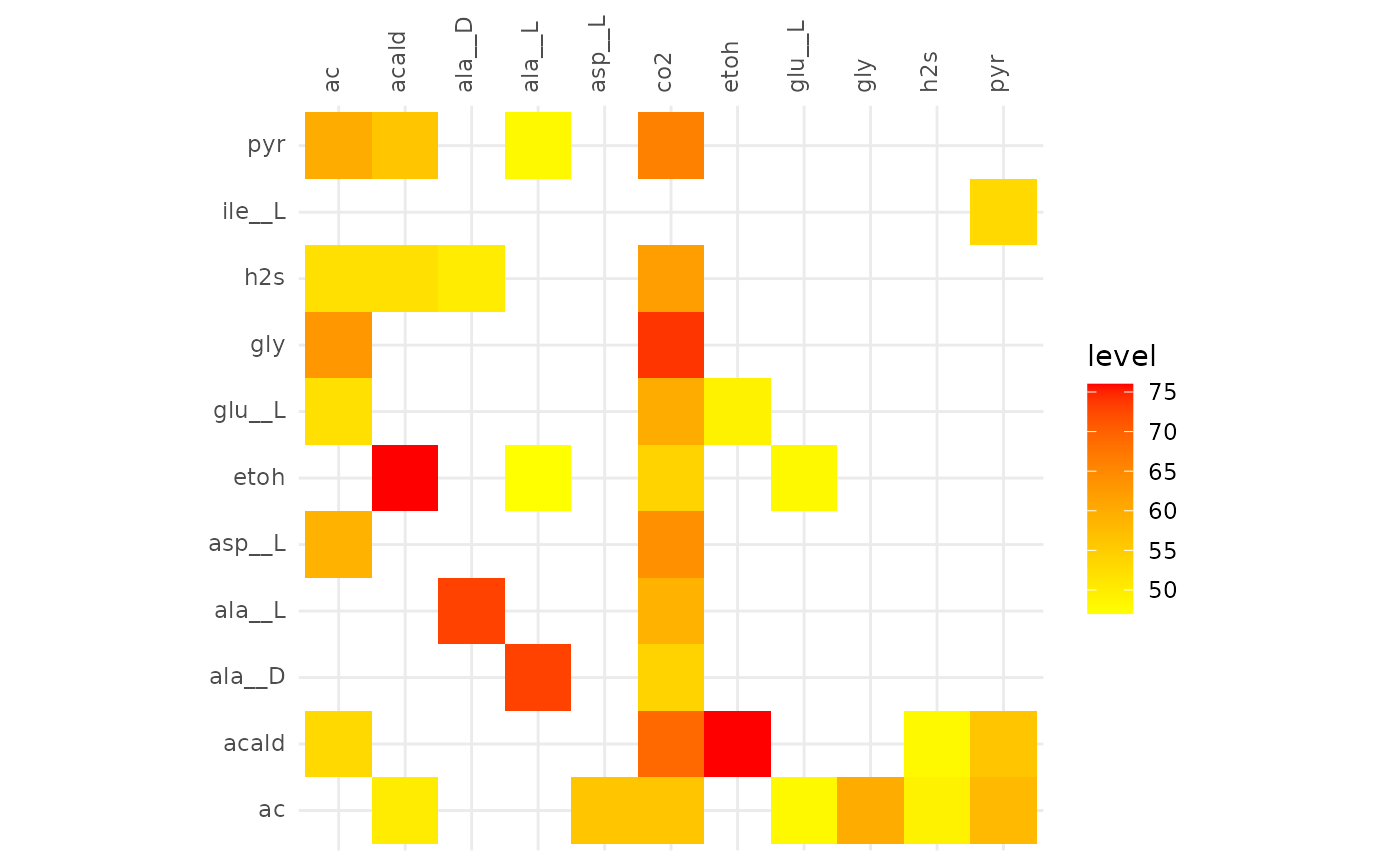
plotAlignmentHeatmap(facilitation_alig, 0.5)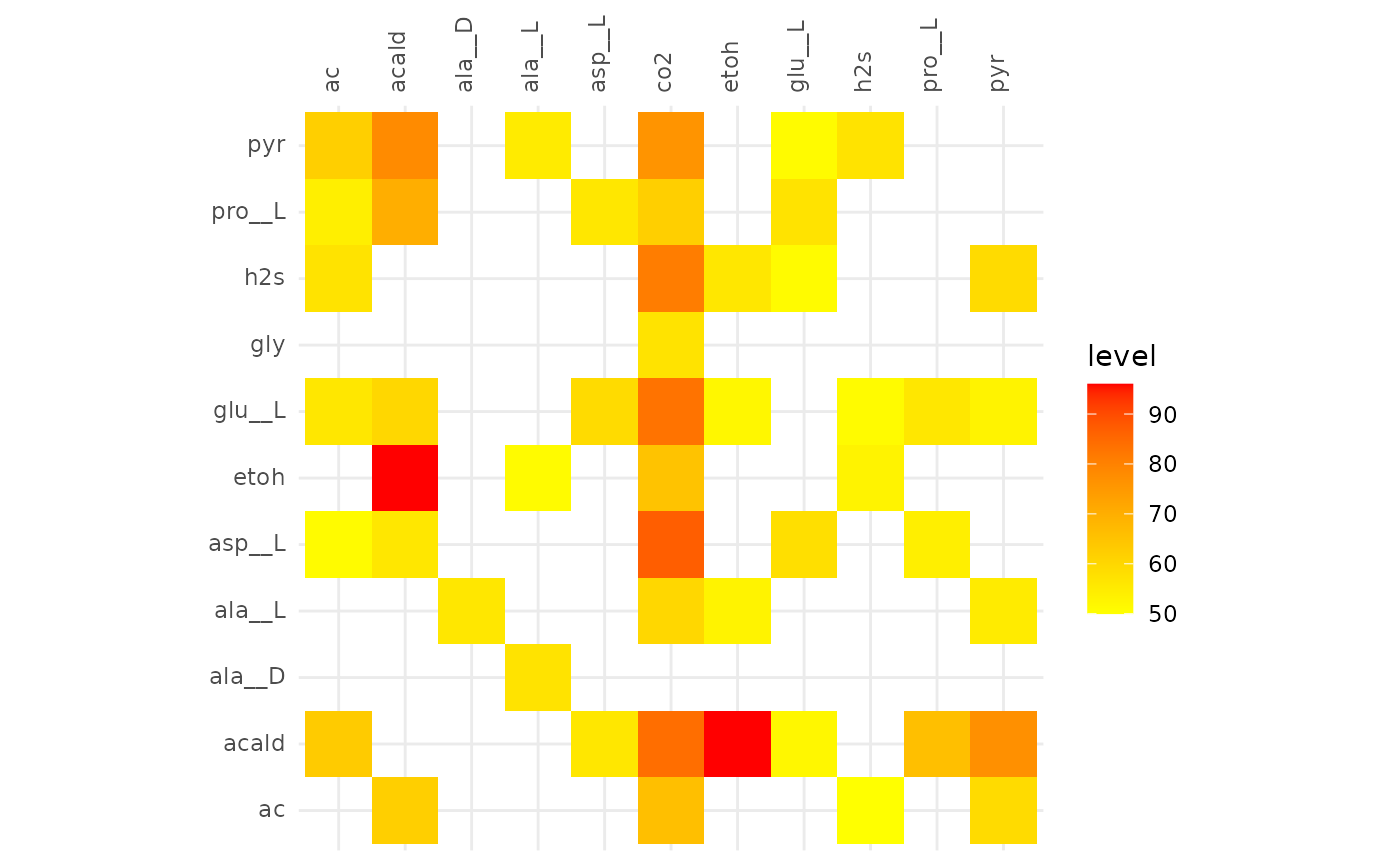
plotAlignmentHeatmap(selection_alig, 0.8)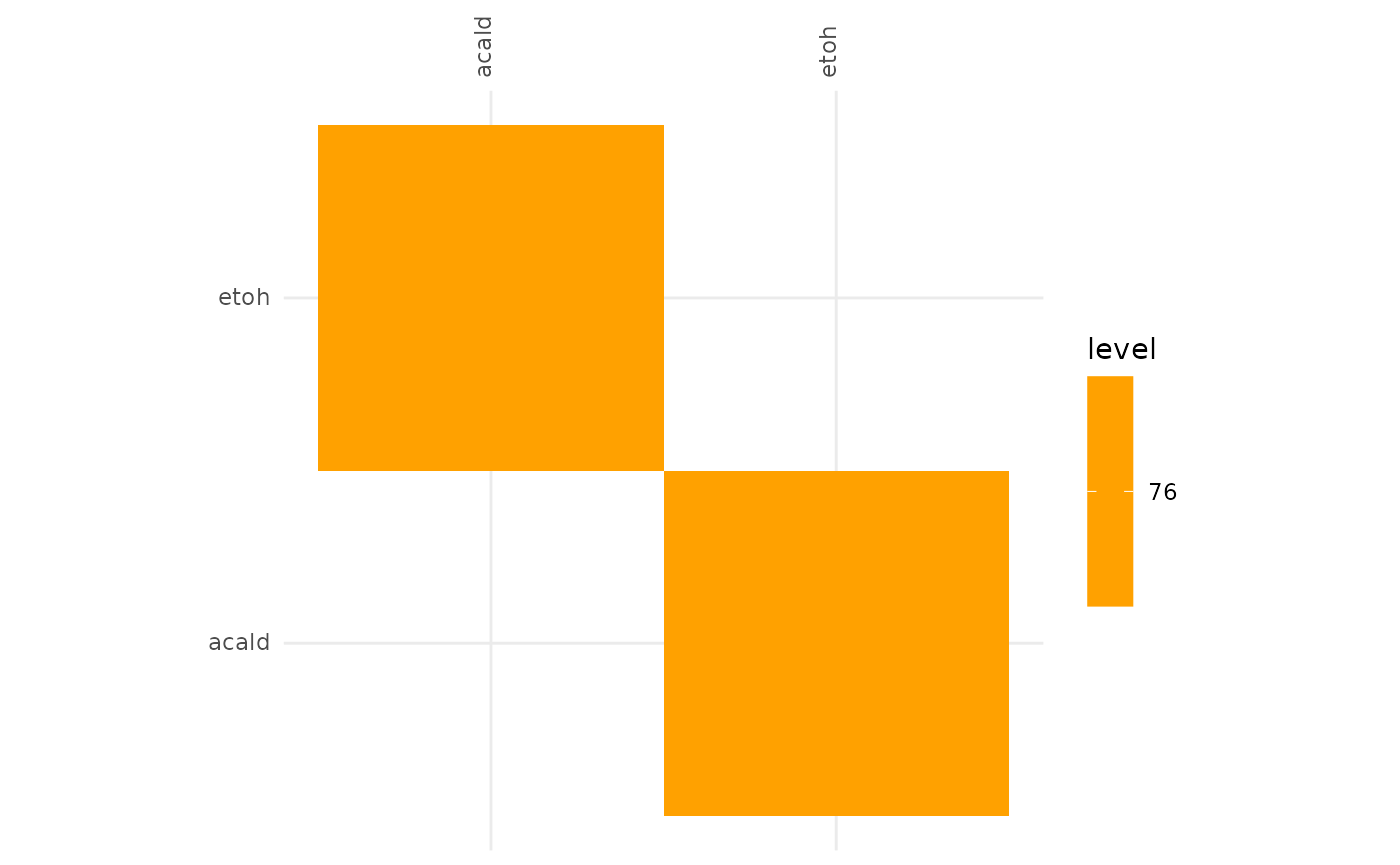
plotAlignmentHeatmap(facilitation_alig, 0.8)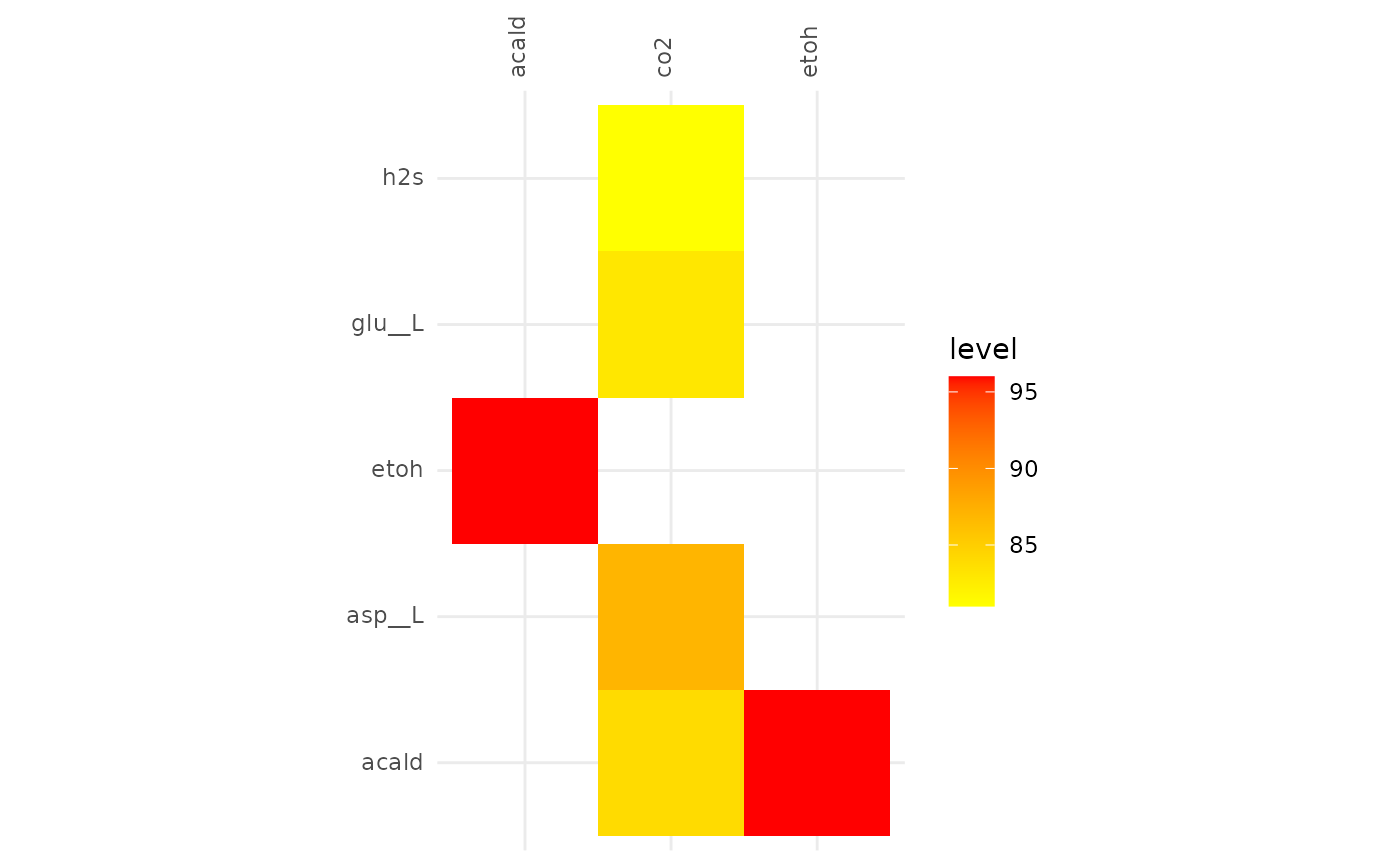
plotAlignmentHeatmap(selection_alig, 0.95)
#> [1] "No Alignment at this level."
plotAlignmentHeatmap(facilitation_alig, 0.95)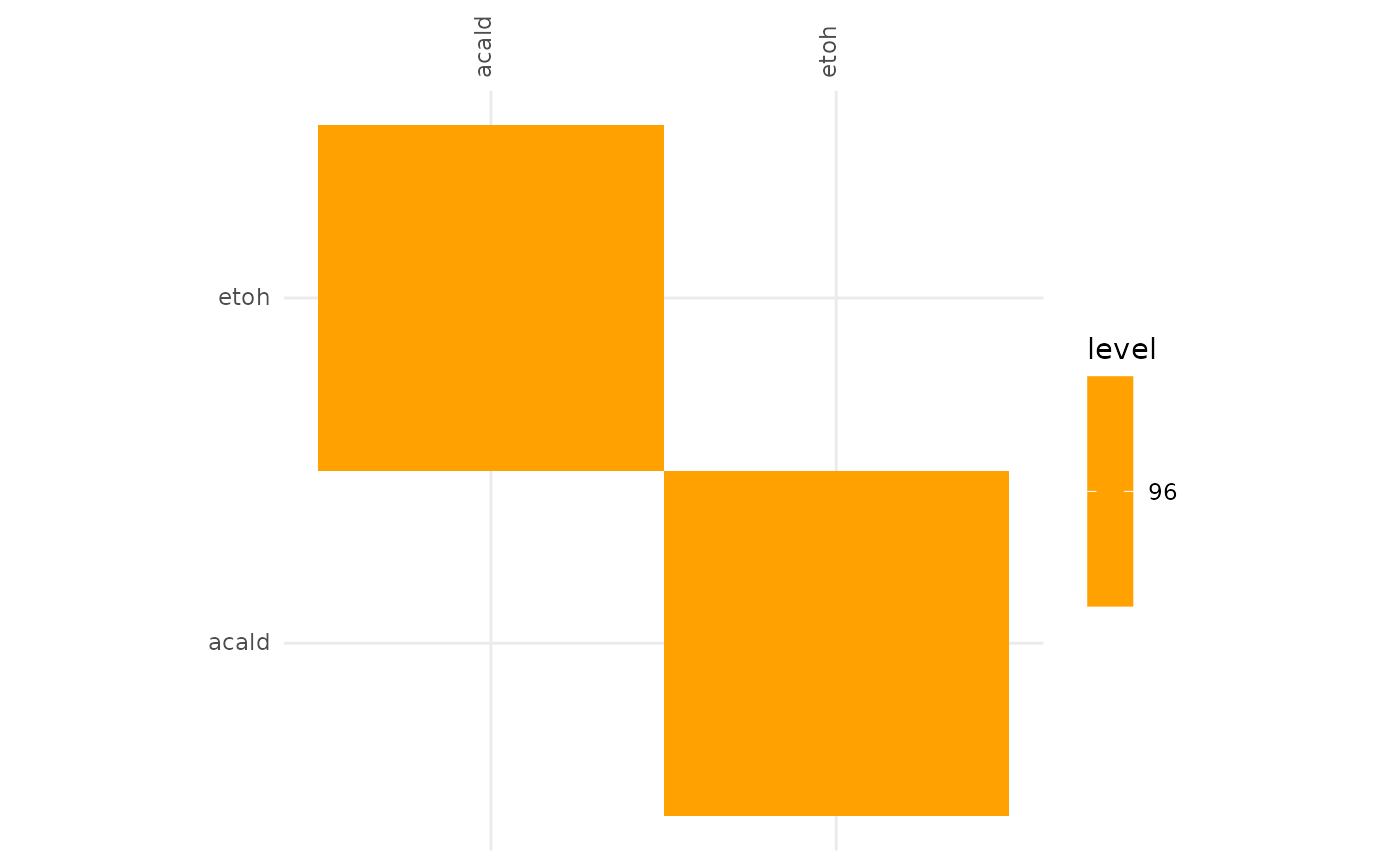
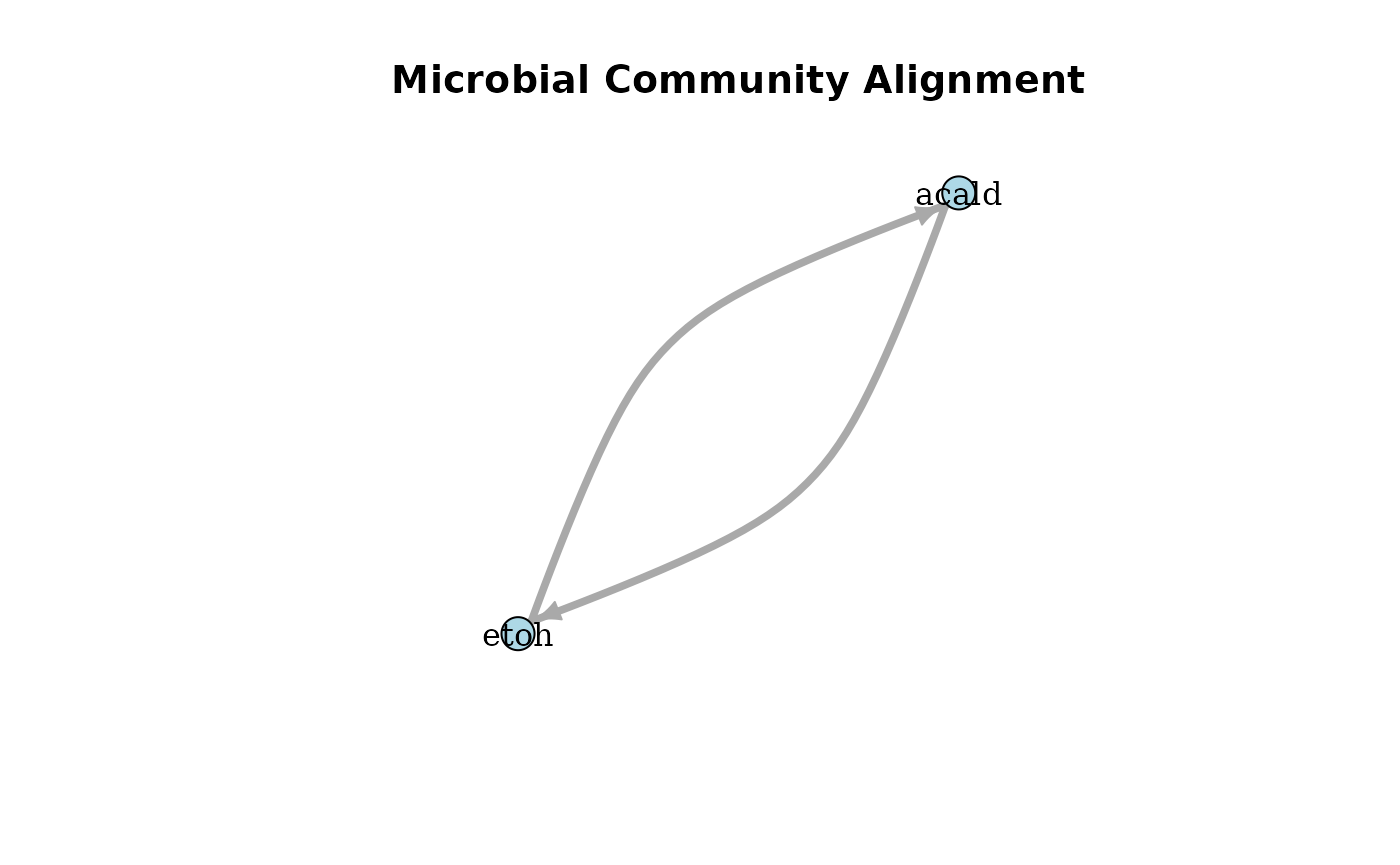
plotAlignmentNetwork(selection_alig, 0.5)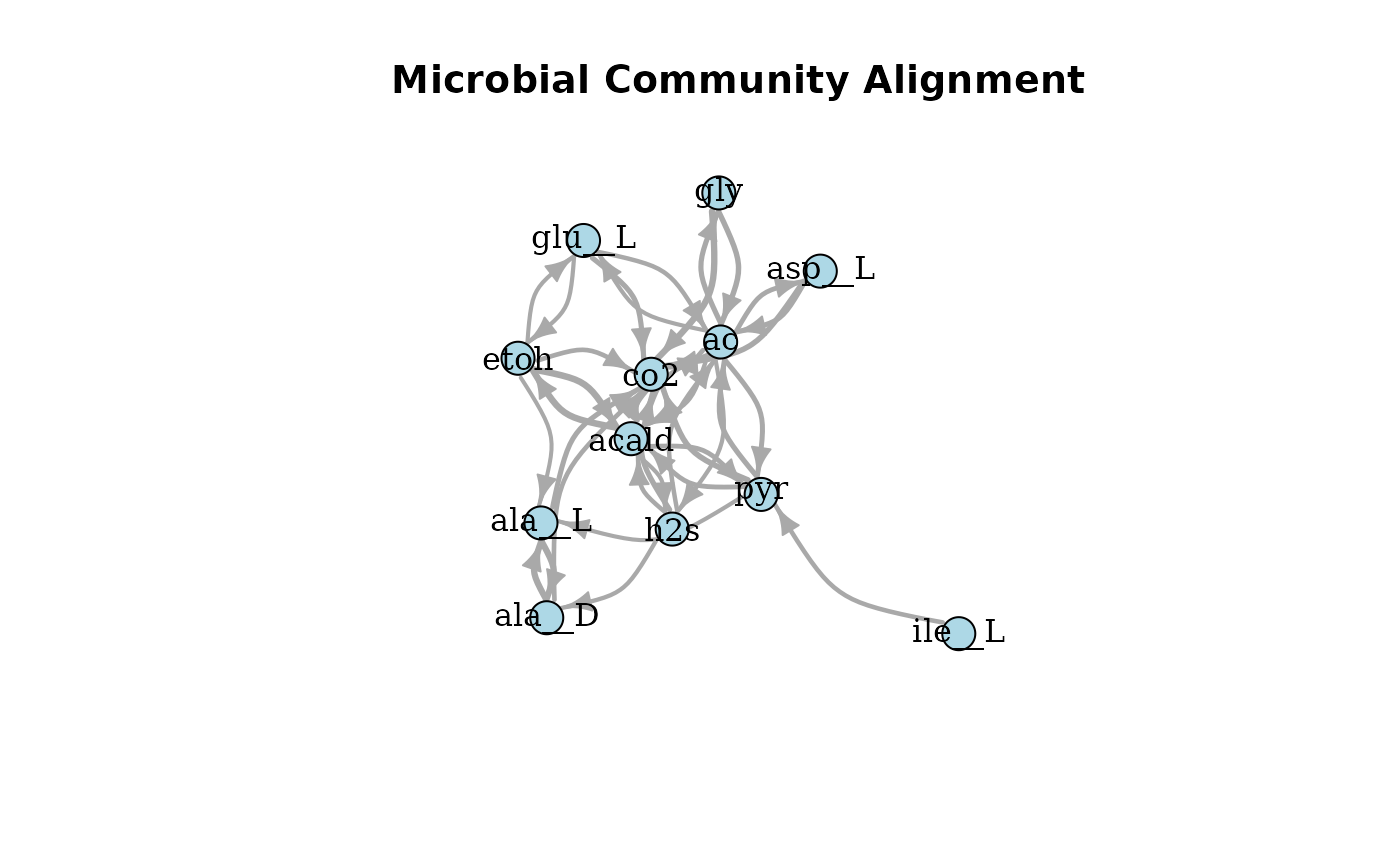
plotAlignmentNetwork(facilitation_alig, 0.5)
plotAlignmentNetwork(selection_alig, 0.8)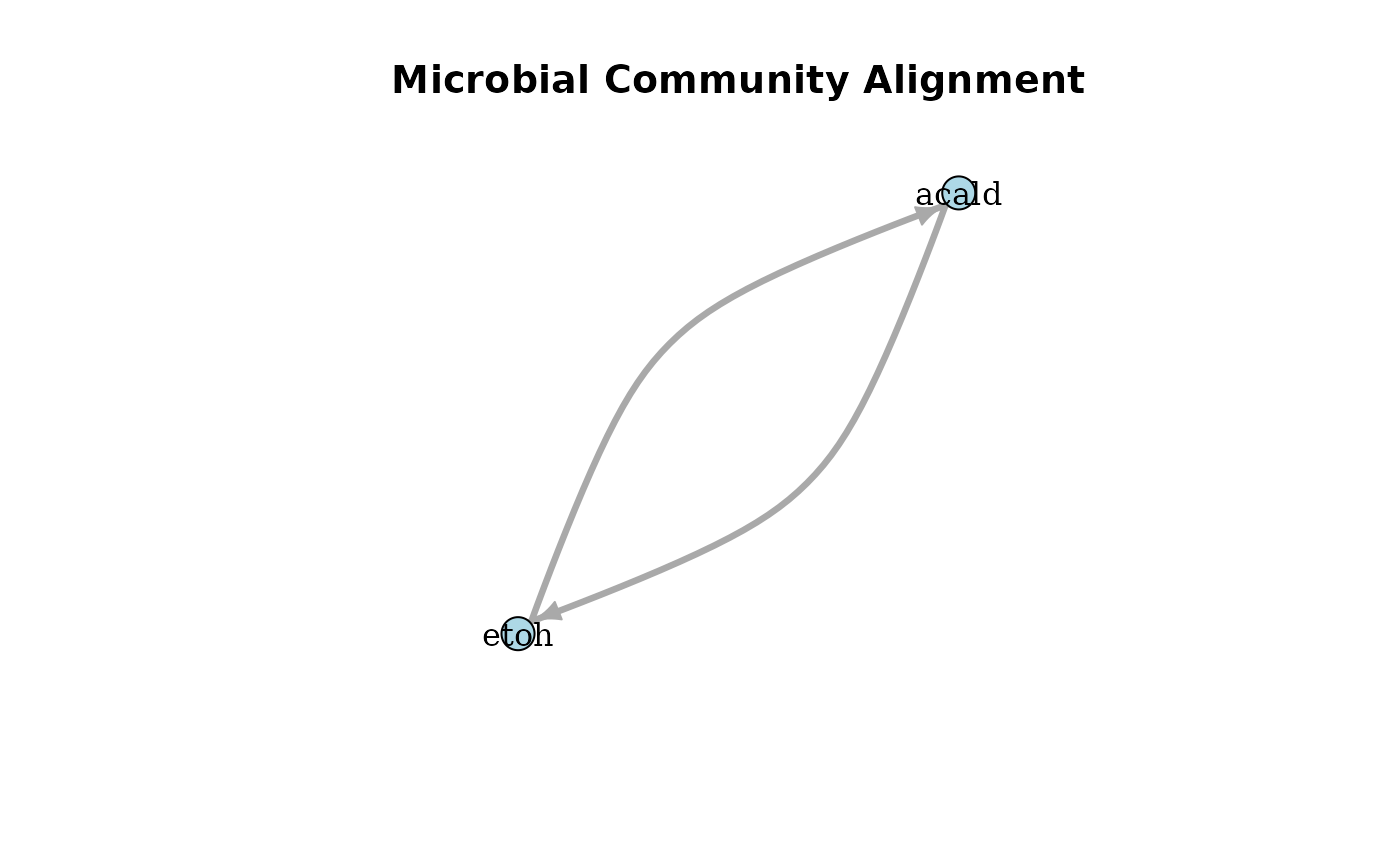
plotAlignmentNetwork(facilitation_alig, 0.8)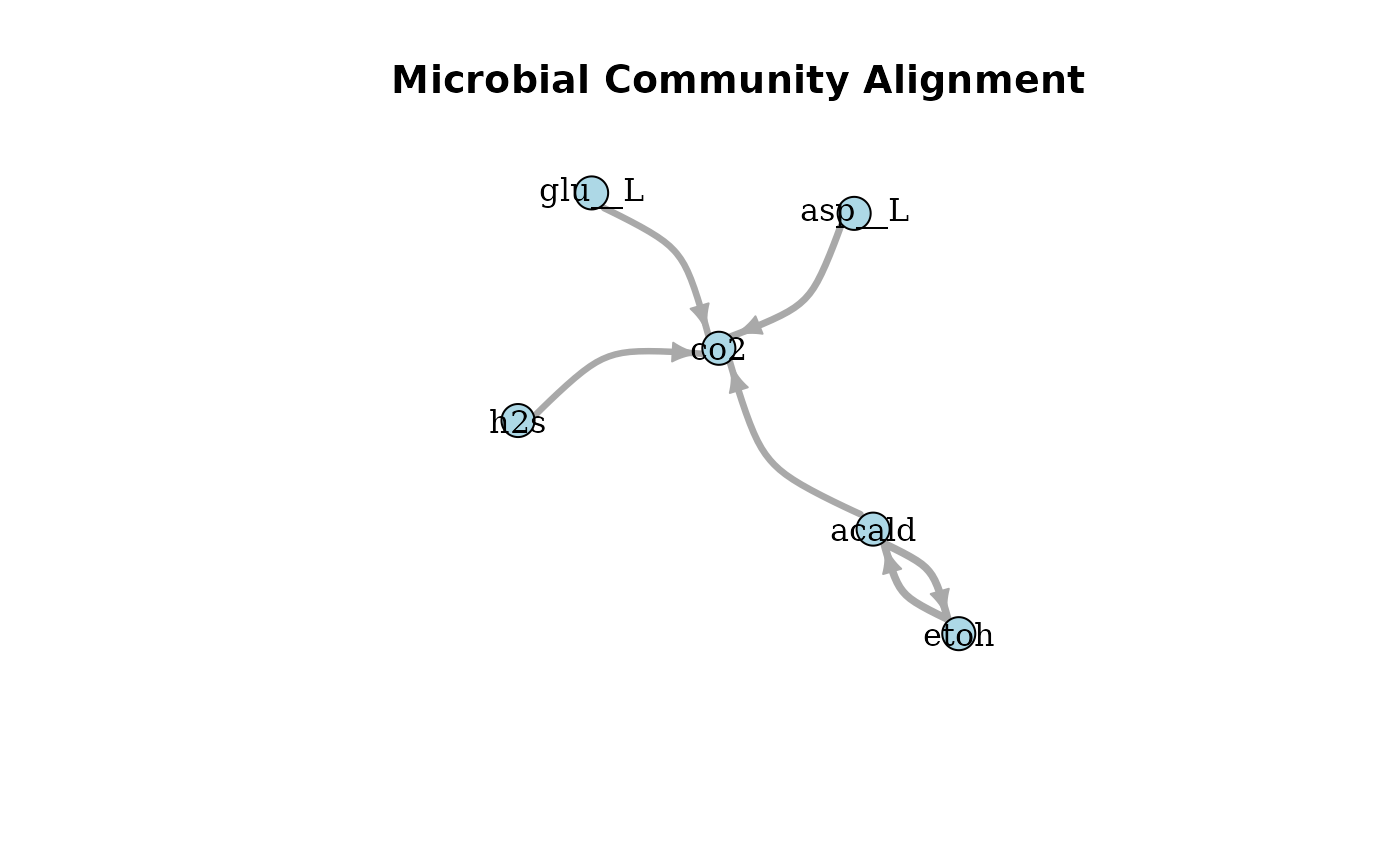
plotAlignmentNetwork(selection_alig, 0.95)
plotAlignmentNetwork(facilitation_alig, 0.95)